Pseudogenes
Typical Protein – By Klaus Hoffmeier – Own work, Public Domain, Link
Pseudogenes have been a hot topic for discussion since their original discovery about fifty years ago. They were originally found in frog DNA and were found to be apparently inactive copies of genes found elsewhere.
They were called “pseudogenes” as they were thought to not make active proteins but represented mutated trash. Thousands of other pseudogenes were subsequently found in humans and many other animals and plants. Many mammals have now been found to have as many pseudogenes as actual protein-coding genes.
Importance in Evolution
Evolutionists jumped on the pseudogenes as showing changes consistent with natural evolution, and inconsistent with intelligent design. They would argue that an “intelligent” designer would not make non-function trash genes. Kenneth Miller called then “discarded sequences” that are
consistent with an evolutionary explanation but inconsistent with intelligent design.

Douglas Futuyma – By Andre levy 72 – Own work, Public Domain, Link
Another evolutionist, Douglas Futuyma wrote that only Darwinian evolution
can explain why the genome is full of ‘fossil’ genes: pseudogenes that have lost their function” that is “hard to reconcile with beneficent intelligent design.
Richard Dawkins, ever the aggressive atheist, wrote
genes that once did something useful but have now been sidelined and are never transcribed or translated. What pseudogenes are useful for is embarrassing creationists. It stretches even their creative ingenuity to make up a convincing reason why an intelligent designer should have created a pseudogene … unless he was deliberately setting out to fool us.
Recent evidence suggests that the optimism posed by the New Atheists may have been premature as they are not functionless.
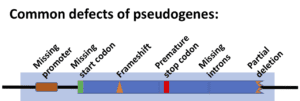
Types of Pseudogenes
There are thought to be three types of pseudogenes:
- Disabled pseudogenes are those sequences of nucleotides that may have once coded for a protein but have been inactivated or rendered useless through mutation,
- Duplicated pseudogenes are multiple copies of still-functioning genes,
- Processed pseudogenes have nucleotide sequences the same as or similar to functioning genes but lack promoter sequences
The majority of pseudogenes fall into the third category in being processed pseudogenes.
Pseudogenes
The 1990s saw a revolution in our understanding of genetics. Scientists discovered that many pseudogenes are still transcribed into RNA. Biologists working with the ENCODE Project sampled 201 pseudogenes and found that at least one-fifth of them are transcribed in one or more tissues.
Some of these RNA molecules resulting from the transcription of pseudogenes produce functional proteins. While only a few pseudogenes produce RNA that is translated into functional proteins, there is growing evidence that these proteins perform essential functions in the cell.
Gene Expression with RNA Interference
Scientists discovered some pseudogenes are transcribed into RNA which might serve to regulate gene expression. The RNA produced is very similar to the RNA produced by a functional gene. The messenger RNA produced by the pseudogene is complementary to the messenger RNA produced by the actual gene, and the two RNAs could bind together. The result would be a double-stranded RNA (- similar to a double-stranded DNA.
The idea that the double-stranded RNA produced by some pseudogenes could suppress the expression of the actual active gene was confirmed in the 1990s in England. An article published in the premier scientific journal Nature in 2008 showed that the RNAs produced from pseudogenes regulate gene expression in mouse eggs. The authors stated their findings
indicate a function for pseudogenes in regulating gene expression by means of the RNA interference pathway.
Gene Expression Enhanced with Pseudogenes
Gene expression might also enhance the expression of the active gene. A group of Japanese and American biologists found that a pseudogene actually enhanced the expression of a mouse enzyme called Makorin-1. They believe this is due to the pseudogene protecting the active gene messenger RNA from degradation.
The hypothesis that the active gene expression might be enhanced by the RNA produced by pseudogenes serving as a false target for degrading enzymes in the cell. In this way, the RNA serves to enhance the gene expression of the active gene through competition for the degrading enzymes.
European biologists then reported the expression of a plant pseudogene increases the expression of a plant gene involved in phosphorus metabolism. Similar to the system described above, the pseudogene produced RNA that serves as a false target for a repressor molecule. These scientists coined the term “target mimicry” to describe the process whereby a pseudogene acts as a false target for a degrading enzyme or a repressor.
Sequence Conservation
Glucose Pathway: By C. Mönchmeier (de:Benutzer:Luziferase) – de:Datei:Gluconeogenese_Schema_2.jpg, CC BY-SA 3.0, Link
Life is very complex, and the unraveling of the interplay between pseudogenes and the actual active genes can be time-consuming and difficult at best. The more scientists unravel these mechanisms, the more other mechanisms become evident.
But there is another way to determine whether an apparently non-functional gene is in fact functional in some unapparent way.
Functional genes are “conserved” meaning they remain the same over time and in different animals if they serve some vital function. This is because variation in the nucleotide sequences in an active gene would likely disturb the functioning of the actual gene product. This means if a gene does not change among very different species, it likely has some undiscovered but important function in the cell’s metabolism.
The ability to rapidly sequence genes over the past few years has made such a determination feasible. Evgeniy Balakirev and Francisco Ayala were able to determine the nucleotide sequences of widely divergent life forms including humans, mice, chickens, and fruit flies. When their data was analyzed, they reported sequence conservation in various pseudogenes – something that was not expected.
If the genes were not important, then pseudogenes in a human might be expected to be different from those found in a fruit fly for example. They found that “pseudogenes are often extremely conserved,” implying the pseudogenes were somehow important to the functioning of the animal and not subject to mutation. Mutations of an important gene would likely alter the gene product in an unfavorable way as most mutations produce loss or reduction of function.
The authors concluded that many genes that are thought to represent “pseudogenes” actually have important functions and are not just “junk DNA.” The idea that many scientists have that DNA is mostly “junk” likely represents ignorance rather than fact.
This finding was duplicated in 2009 by a group of Canadian biologists who evaluated genes in humans, monkeys, mice, rats, dogs, and cows. They found significant sequence similarity among these “pseudogenes” suggesting they had been conserved for a purpose not yet known. They concluded,
through evolutionary analysis, we have identified candidate sequences for functional human transcribed pseudogenes.
Summary
Pseudogenes have been displayed as an important supporting finding for Darwinian evolution. The finding of non-functional copies of functional genes would lend support to modern evolutionary theory rather than intelligent design.
Various evolutionary biologists including Richard Dawkins, Douglas Futuyma, Michael Shermer, Jerry Coyne, and John Avise all have argued that pseudogenes support Darwinism because they are non-functional. Even more important, they suggest that an intelligent designer would not make genes that have no discernable function.
Now we know their optimism has been overplayed. Rather than supporting Darwinism, more detailed analysis suggests these “pseudogenes” may have a function. The finding that the gene sequence is conserved among animals as divergent as humans and fruit flies implies some undiscovered function for these genes making the case for an intelligent creator.
Further research will likely uncover functionality for many genes which have been thought to have no function. This would be the reason why gene structure has remained the same or very similar with genes shared among such divergent forms of life.